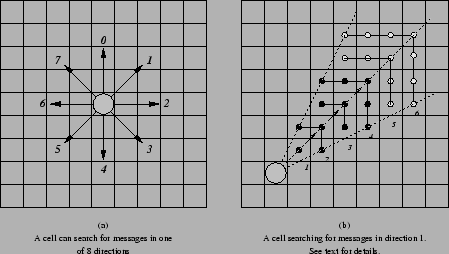
 |
A cell can issue a rms_receive instruction to receive messages which have been broadcast from nearby grid positions. These messages (which are EnvironmentalInfoStrings), like all InfoStrings, have a type (a number between 0 and 15) associated with them, and the value of a cell's dx register at the time that it issues a rms_receive specifies which type of messages are to be received. In addition, the search in the environment for EnvironmentalInfoStrings of the specified type only proceeds in a certain direction; starting from the grid position of the cell that issued the instruction, the search emanates in one of eight directions, specified by the low three bits of the cx register (see Figure 4.3(a)). The search proceeds one grid square at a time, covering all grid squares in the specified eighth of the area around the cell until a certain number of grid squares have been searched (defined by the global parameter rms_receive_search_area). For example, Figure 4.3(b) shows a cell searching in direction 1. If rms_receive_search_area is set to 12, say, then the grid positions marked with black dots will be searched. The search emanates from the cell along a series of wavefronts--the grid position on wavefront 1 is searched first, followed by those on wavefront 2, then 3, then 4. At this point, 12 positions have been visited, so the search stops. Any EnvironmentalInfoStrings of the specified type found in this area are copied into the cell's Received Message Store as InfoStrings. (A cell may extend the reach of a search by re-issuing an identical rms_receive instruction from the same grid position within a certain time limit after the first one. This time limit is specified by the global parameter max_time_for_msg_receive_reinforcement. If a cell does this, the search will continue outwards from the last grid position searched previously. In the example of Figure 4.3(b), the grid positions marked with gray dots, on wavefronts 5 and 6, will be the next 12 positions searched in this situation.)
The host cell may process these received messages, using the str_switch and adr instructions to set the ax register to an address within a message, and using the instruction mov_ic to sequentially read the message.
Messages in the Received Message Store are normally treated as passive structures which may be inspected by the host code, but this is not always the case. As already mentioned, each message in the store has an associated type. The host code of the cell--the genome--being an InfoString, also has a type associated with it.4.12 If any message in the Received Message Store happens to be of the same InfoString type as the cell's genome, then it may potentially be used as additional genetic material, and translated into executable instructions. In other words, promoters and repressors may bind to it in just the same way as they can bind to the genome. If the active promoter does indeed bind to a message in the Received Message Store, translation begins along it just as it would on the genome. A cell has several lines of defence against such parasitism, which are mentioned in Section 4.6.1.
A situation where the execution of code from messages in the Received Message Store may be particularly common is when the parameter neighbouring_genomes_readable is set to yes. In this case, whenever a new promoter becomes active in the cell (see Section 4.3.3), rather than trying to first find a binding site on the cell's genome, or even on eligible messages already resident in the Received Message Store, the cell first imports copies of the genomes of any immediately neighbouring cells, one by one, into its Received Message Store. Note that this importation occurs automatically, without the host cell having to issue a rms_receive instruction, and without the neighbouring cell having to make a copy of its genome and issue a cwm_send instruction. Each imported message (the copy of the neighbouring cell's genome) is checked for a binding site for the new promoter. If a site is found, the message remains in the Received Message Store, and the cell starts executing instructions from it, starting in the position immediately following the binding site (see Section 4.3.3). If no binding site is found, the cell deletes the imported message from its Received Message Store and imports the genome of the next neighbouring cell, if there are any remaining. Only after all the neighbouring genomes have been checked in this way will the cell consider searching for a binding site on existing messages in the Received Message Store, and finally on the cell's genome itself. This mechanism was incorporated into the system in an effort to simulate the ability of programs in Tierra to read the code of neighbouring programs [Ray 91].