 |
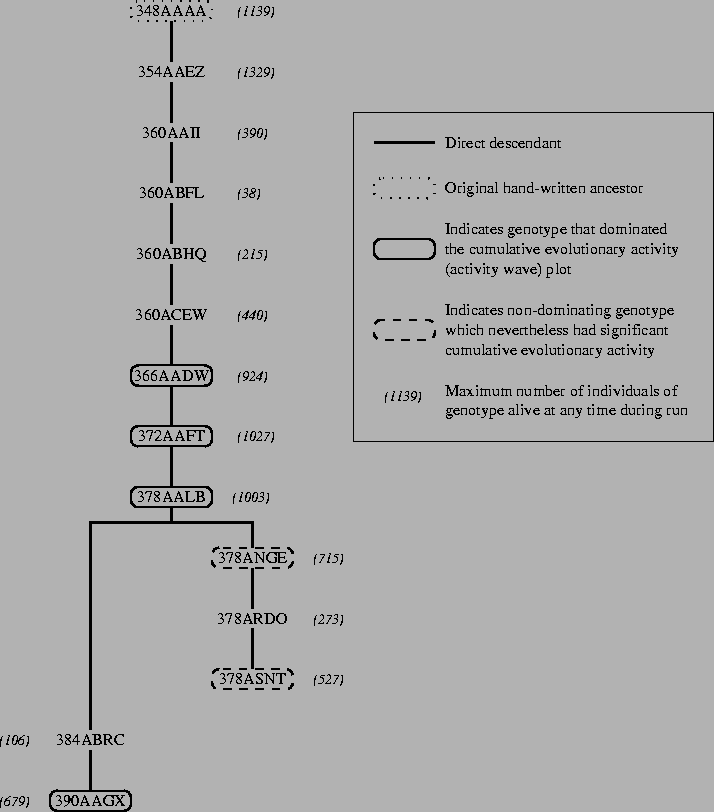
By analysing the output data from the run, it is possible to reconstruct the phylogenetic tree (the evolutionary history) of the significant genotypes during the run. The reconstruction for the standard run is shown in Figure 5.24.
An interesting point to note about the phylogeny is that some of the ancestors of the genotypes that were significant throughout the course of the run did not themselves enjoy much success. An extreme example is genotype 360ABFL, from which all of the subsequent adaptively significant genotypes are descended. Despite its significance as an ancestor, the number of programs of genotype 360ABFL existing at any one time was never more than 38 (in a total population of over 800), and the genotype only existed for about 4000 time slices in total. Had circumstances been slightly different, the genotype might have died off before giving rise to its descendant 360ABHQ, or indeed 360ABFL might never have arisen at all. This indicates that the role of chance might play a large part in determining the outcome of a single evolutionary run. We will look into this issue in detail in Section 6.1.
Each row presents data on the comparison of a genotype (first column) with its ancestor (second column). Columns 3-8 refer to subsections of the genome (c.f. Figure 5.1; the subsections labelled begin_template and init have been grouped into a section labelled begin here, and similarly the subsections move, stop and end_template have been grouped together as end). In the row below the subsection name is the length of that subsection (in number of instructions) in the original ancestor 348AAAA. The column marked + shows how many new instructions were added to this subsection during the evolution from the indicated ancestor. The column marked - shows how many instructions were removed. Numbers in braces () in the + column show how many of the added instructions were the energy collection instruction (et_collect). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
A summary of the sources of innovation from ancestor to descendent for the most significant genotypes is presented in Table 5.2. The table reveals that the only sections of the genome which vary between the genotypes are the copy_loop and end sections.
Looking at the changes in the copy_loop section, we see that all of the additions were extra et_collect instructions. Comparing the final dominant genotype 390AAGX with the original hand-written ancestor 348AAAA, seven extra et_collect instructions have been added within the copy_loop section. If we compare the other subsection that experienced changes, the end section, we see that it only actually grew by one instruction from 348AAAA to 390AAGX (there were three additions and two removals). These findings therefore confirm the prediction made in Sections 5.2.2 and 5.2.4 that the programs are evolving to collect more energy, and that the extra energy collection instructions are being placed within the copy loop, so that they are executed at each iteration of the loop.
The changes made in the end section included the deletion of the move instruction in genotype 366AADW (the genome of the ancestor 348AAAA is shown in Section B.2). None of the subsequent dominant genotypes re-inserted this instruction, so the ability to move around the grid apparently did not convey an adaptive advantage in this run. The stop instruction was also deleted (both this and the move instruction were replaced by instructions which had no effect in the program), with subsequent genotypes taking advantage of the fact that when program execution reached the final instruction in the genome, the stop instruction was implicitly executed anyway. The extra instruction added at the end of the end section (in the transition from genotype 348AAAA to 366AADW) was an extra copy of the final instruction in the program (which actually formed part of the end template). This extra instruction appeared as a consequence of the if_not_fl instruction in the copy loop being replaced by an et_collect. The removal of the if_not_fl meant that the program did not check whether it had already reached the position marked by the end template before copying the next instruction to its offspring, so the final section of the end template was copied twice before the copy loop was exited. Apparently, the inclusion of an extra et_collect in the copy loop outweighed the penalty of having an extra, redundant instruction at the end of the program.