 |
As mentioned in the previous chapter, there are many parameters associated with Cosmos, some of which control fairly minor or obscure aspects of the system. Because of this, it would be impractical to conduct a thorough investigation of the entire parameter space. Instead, many of the more obscure parameters were held constant throughout all of the experiments reported in this chapter. These are listed in Section B.1, along with their associated values.
The parameters which were varied between experiments (together with some other major parameters such as grid_size) are listed in Table 5.1. This table also shows the default values of these parameters, as used in the run reported in this section.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
As shown in the table, the run was initialised by inoculating 100 ancestor programs into the system. The size of the world was 40 x 40 squares, and the ancestors were distributed evenly throughout this area.
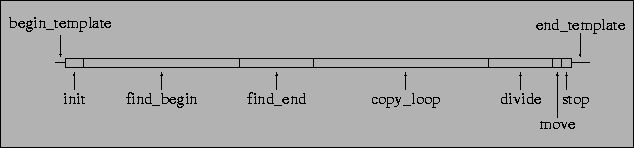
The ancestor used for this run (and for nearly all of the other runs) is shown schematically in Figure 5.1 (a full listing is given in Section B.2). The basic operation of the ancestor is as follows. First, it looks for a template pattern at the start of its genome to work out the memory address of its first line of code. Next, it searches for a template pattern at the end of the genome, to work out the address of its last line of code. The ancestor then enters into a loop, copying instructions one at a time, from the beginning of the genome to the end, into the Nucleus Working Memory. When this copying is complete, the cells divides (creating a new organism), and the ancestor attempts to move into a neighbouring grid position.
The duration of the run was one million time slices. As discussed in Section 5.1.4, data for the neutral shadow was recorded during this run, and then fed into the neutral model to give us an idea of the non-adaptive characteristics of the system.